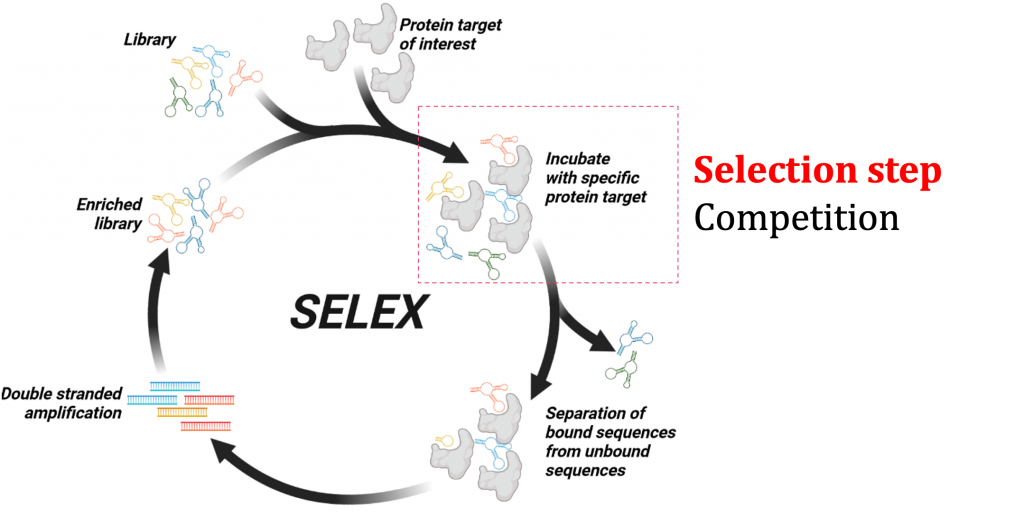
Nucleic Acid Aptamers
We are working with a special class of nucleic acid molecules called aptamers. We want to develop biochemical tools using aptamers against membrane proteins to understand how cell receptors respond to external stimuli. Aptamers are short and single-stranded DNA/RNA sequences that specifically bind to a target molecule with high affinity. Aptamer binding is based on the ability of DNA or RNA (typically 40-100mers) to fold into unique three-dimensional structures and their ability to interact with a binding epitope of the target molecule. Aptamers show some inherent advantages that merit their application in biomedical sciences. One fundamental advantage of aptamers is the ability to introduce chemistries that allow proper quality control. Aptamers are also smaller in size. The smaller size also contributes to higher clearance rates. The synthetic nature and small size will also be beneficial in designing better molecules for therapy, diagnostic, and biochemical tools. The selection process of aptamers is called the Systematic Evolution of Ligands by EXponential enrichment (SELEX). Typically, the SELEX process combines in vitro evolution and combinatorial chemistry. Aptamers selected using traditional SELEX methods using purified membrane proteins in solution have shown limited translational applicability.
Ligand Guided Selection
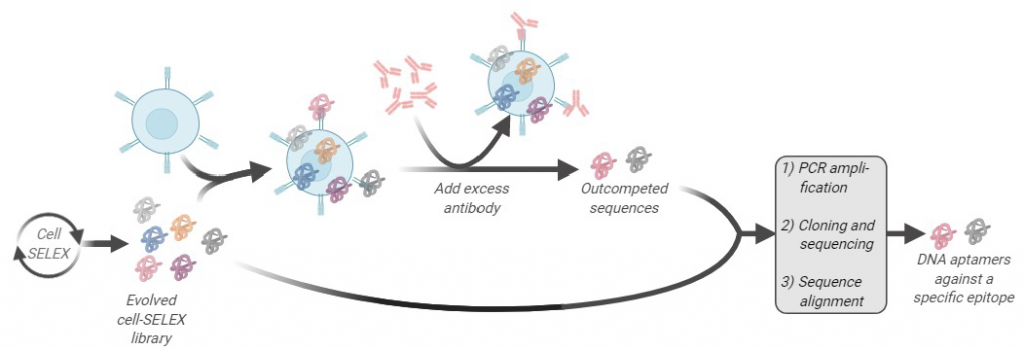
One of the major research avenues in our lab is to develop new aptamer screening technologies using the SELEX platform. Through these new screening methods, we aim to understand the evolution of an aptamer library while at the same time identifying aptamers with better binding properties against membrane proteins. We recently pioneered a method called Ligand Guided Selection (LIGS). One central theme of our research is that a pre-existing interaction of a receptor and a ligand can be exploited to identify artificial ligands from a library of synthetic molecules against the same receptor the natural ligand binds to. The aptamer ligands identified using LIGS show higher specificity, allowing us to engineer molecular tools against cell membrane proteins.
In a combinatorial library of molecules, the concentration of the artificial ligand is limited, and by adding an excess amount of a pre-existing naturally occurring ligand, the following can be achieved:
- To selectively out-compete the synthetic ligand that is competing with the natural ligand to bind to the same protein
- To induce a conformational switch of the protein so that the binding site of the protein is no longer desirable towards the artificial ligand, resulting in the elution of the artificial ligand.
Our prototype-screening library is an evolved nucleic acid ligand library against the target of interest. We used an antibody interacting with its antigen as the pre-existing receptor-ligand interaction to show this concept. Using an evolved SELEX library against a pre-determined receptor protein expressed on the cell surface, in conjunction with respective antibodies interacting with the same receptor, we showed the functional selection of specific aptamers against a specific epitope of a receptor protein could be done. We call this technology “Ligand Guided Selection (LIGS),” and it is important to note that LIGS can be expanded to identify other types of artificial ligands, such as peptide-aptamers, small synthetic ligands, Fab fragments, toward a protein that have cognate ligand, from their respective libraries.
Molecular Engineering and the Synthesis of Nucleic Acid Analogs
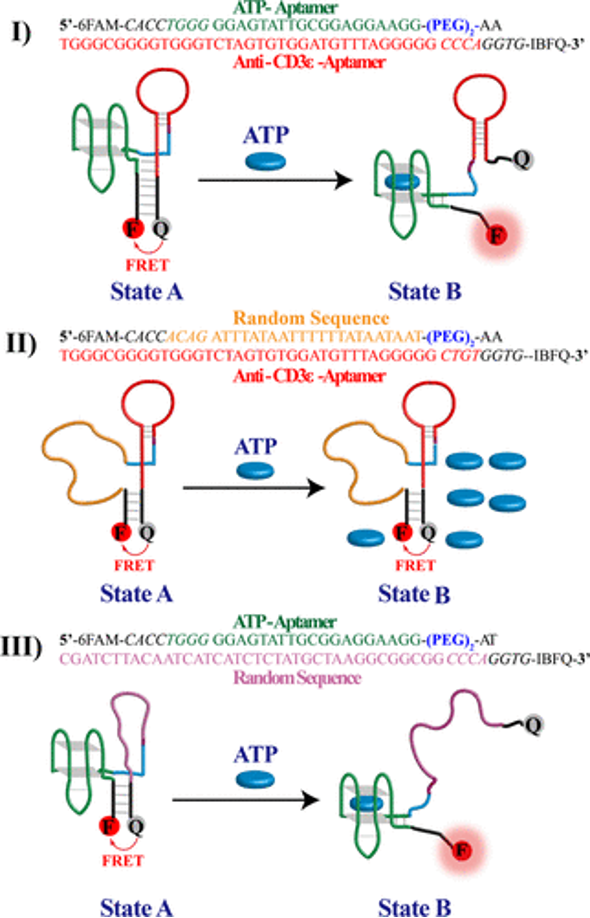
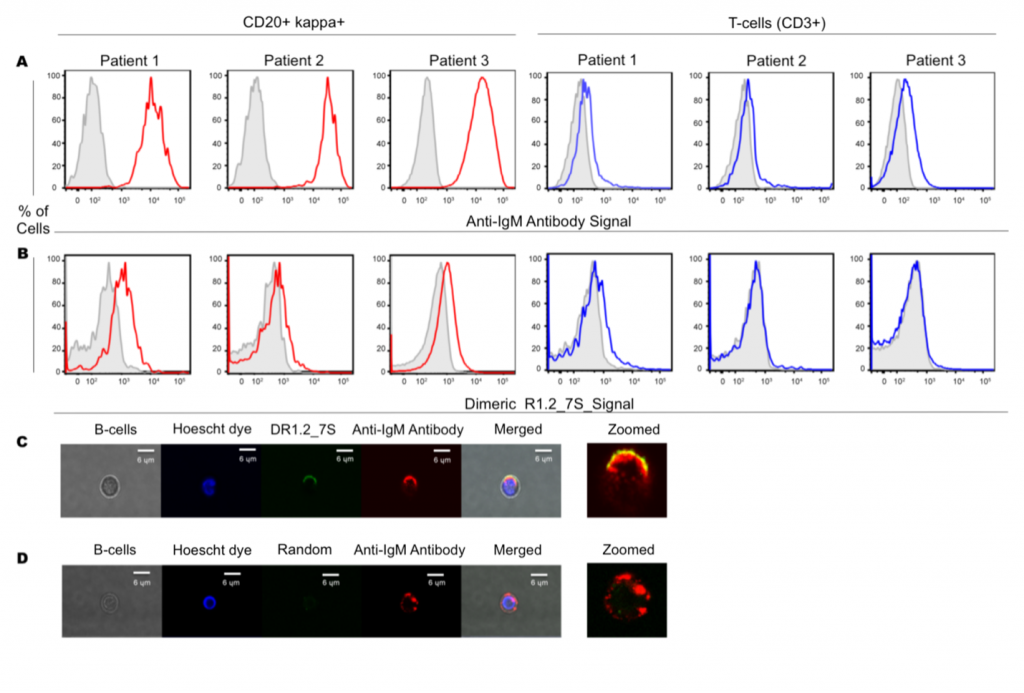
Our primary focuses are developing bispecific aptamers for immunomodulation, bispecific aptamers with superior sensing capabilities, and bivalent aptamers as ligands against cell surface receptors. We draw inspiration from naturally occurring molecular scaffolds to engineer new molecules based on aptamers. These molecules are then used to tackle and understand the cytolytic responses of the immune system. We recently introduced bivalent aptamers capable of activating TCR-CD3 receptors and bivalent aptamers with higher affinity capable of binding to its targets in primary lymphoma cells. We are synthesizing nucleic analogs to find a way to improve the chemical space of nucleic acid aptamers. One of the focuses is to make triphosphate and phosphoramidites to be apply in designing new ligands and molecular tools with higher chemical space.
If you want further details about the work, please read manuscripts and review articles listed under publications.
